
Other automated products include ACT, DARWIN, DNASIS GeneIndex, Entrez Cross Database Searcher, PatternHunter, Seqware DataCenter, and StarBlast. It is hardly alone, however, in providing database access.īaylor College of Medicines SearchLauncher (listed as BLAST) is one of the best, but there are zillions of others not listed here for space reasons.
#Macvector protein sequence software#
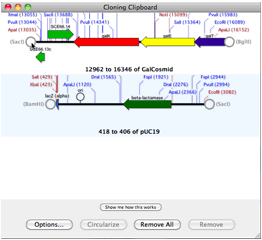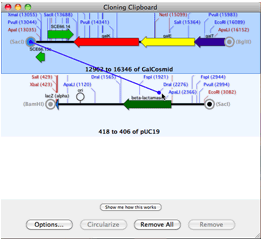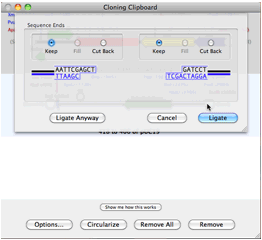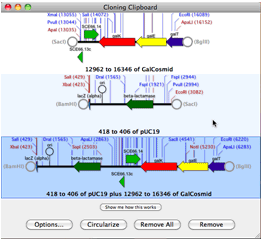
At least one integrated software sequence editor (MacVector) has stood out, providing users with the simplest interface for accessing Entrez. In some ways, this category overlaps with sequence editors. Sequence searching, as a category, has a considerable hodge-podge of offerings. Notably, all of the products listed in this category are noncommercial, reflecting the fact that commercial software developers have looked to more complicated translation problems, such as gene identification in eukaryotes (genomics), as posing more interesting problems and having more potential for development. Numerous tools are available for simple identification and translation of protein coding sequences in DNA. Researchers demands for publication-quality graphic products have fueled development of plasmid mapping software (Gene Construction Kit, NetPlasmid, PDRAW32, REMAP, SimVector, and Visual Cloning) and it is this function that has seen considerable development. Though identification of restriction sites from sequences is still important for vector construction, online tools (In Silico Restriction Cutting, NEB Cutter, WatCut), freebies (EnzymeX), and integrated products mentioned above satisfy these needs well. Restriction enzyme related software is another category whose original usefulness has paled in comparison to bigger project demands and has expanded to meet new needs. Offerings here include CAP3, iCE, MERGER, MIRA2, Paracel Genome Assembler, Paracel Transcript Assembler, and Sequencher.Ī few tools for sequence manipulation, such as translating, reverse complementing, and inverting are still out there (Reverse Complement), but this subcategory serves as little more than a reminder of computational problems of long ago.
The third is for assembly of contigs generated by sequencing projects. The second is the development of software to read automated sequence information (4Peaks, ABI View, PHRED, TraceTuner). A few products meet those needs (biOpen, CINEMA, ElDorado, STING). The first is the desire of users to format sequences with color, annotations, and other information. The category of sequence display/editing/manipulation has expanded considerably in recent years as demands peripheral to simple sequence editing have arisen from genome projects. MacVector users rejoiced in the release of a new version (8.0) designed to take advantage of OS X, breathing life into Bill Kraus original standard that keeps plugging along. The past couple of years have witnessed one commercial product resurrected from the dead (DNASIS), and one collection suffering from neglect (ChromaTool, GeneTool, and PepTool), as well as expansion (LaserGene), and new ownership (Vector NTI Suite, acquired by Invitrogen). Despite inroads being made by the open source movement, commercial vendors appear to be holding on, at least as measured by their numbers. The category of integrated software products has grown in number with a couple of new additions (DNAssist and Sequence Manipulation Suite). Overall sequence similarity with the mouse protein is shown on the right.Microarrays have provided very fertile programming problems, with software products that assist in almost every aspect of array-related research, from design to visualization to statistical analysis to integration of information to system-wide understanding. The percent similarity of the SAM domains and other regions to the corresponding regions of the mouse protein is shown. (D) Schematic comparison of the amino acid sequences for mouse, rat, human, chick and zebrafish mr-s proteins. Branch lengths reflect the mean number of substitutions per site. Amino acid sequences were analyzed by the neighbor-joining method in MacVector 7.2. (C) Phylogenetic tree of SAM domain-containing proteins. The sites that were targeted for mutagenesis are indicated by arrows. Conserved amino acid residues are shown with a dark shadow and functionally similar residues are shown with a light shadow. (B) Alignment of SAM domain sequences for SAM domain-containing proteins. The underline indicates a putative polyadenylation termination signal. Boxed amino acids are the SAM domain sequence and the dashed box indicates a putative nuclear localization signal. (A) mr-s nucleotide and amino acids sequences. Mr-s nucleotide and amino acid sequences.


 0 kommentar(er)
0 kommentar(er)
